:dizzy:It would make sense for them to either all be the same or all be different. But we find neither
So when you design a lot of different things, they have to either be all the same or all different. That's your argument?
Calling me names was more convincing.
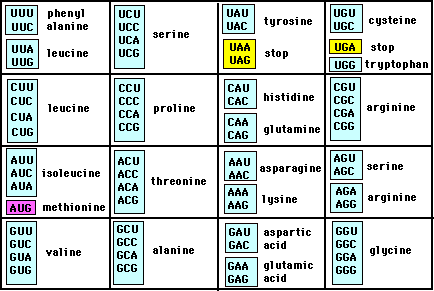
You know the answer to this. :thumb:WHY are they so similar Stripe? Why are whales closer to cows than any other mammal in that listing of creatures?
Of course it's scientific to point to the truth!Not a scientific argument.
Evolutionists love to assume the truth of that which they hold dear and use it for evidence.You have specific ideas about HOW God created. Saying "God just did it that way" is a total cop out. You're then saying God made it look like evolution without actually creating through evolution.
:yawn:You mean all of them? I'm asking for YOU to use YOUR definition. I can give you examples of species, and why they are defined as such all day. If "kind" is so simple and obvious, you should have no trouble giving us examples ad nauseum.
Feel free to not talk to me. :thumb:All you've done in this entire thread is assert, obfuscate and dodge. You're a total waste of time.